Pandemic year in review
What a journey! Today marks one year from when I last did an in-person seminar – to the Medzhitov Lab at Yale University in a giant socially-distanced room (see inset photo). We could all sense the impending changes, but could not have fathomed the extent to which they would transform society.

To bring into focus my growing lab group at Arizona State University, including how our trajectory has broadened during the pandemic, let’s trace the rough milestones of the past year leading up to the present moment:
-
March 2020: Reality of a global pandemic strikes; lockdown contemplations about non-linear risk led to this coronavirus vs. climate change post on Medium co-authored with the awesome Liliana Dávalos.
-
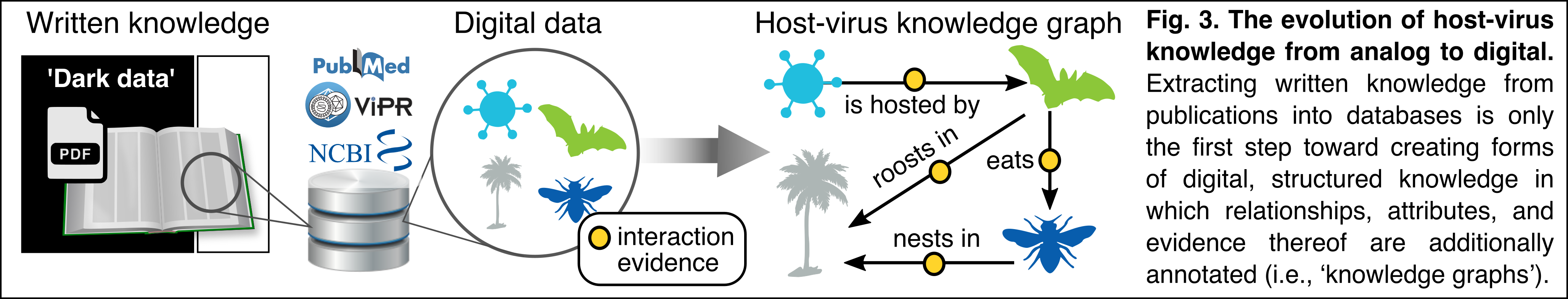
April-July 2020: Wanting to learn how mammal biodiversity & natural history data could help prevent future zoonotic outbreaks, I joined a Covid-19 Taskforce of museum-based biodiversity scientists from the US, Europe, and Brazil (organized by iDigBio, Consortium of European Taxonomic Facilities [CETAF], and the Distributed System of Scientific Collections [DiSSCo]). Via a series of weekly Zoom sessions and break-out groups, I connected with a dynamic group of researchers from Plazi (Donat Agosti), GloBI (Jorrit Poelen), and Pensoft Publishers (Lyubo Penev), among many others. These discussions coalesced around the need to freely connect specimen-based biodiversity information, which is often locked in disconnected publications, into a broad ‘Hub’ of mammal host-to-virus data. Having connected digital knowledge should allow for much higher resolution estimates of taxon-specific zoonotic risk than current cyberinfrastructures allows. Outputs from this work include:
-
Policy article pre-print. Upham, N. S., Poelen, J. H., Paul, D. L., Groom, Q. J., Simmons, N. B., Vanhove, M. P. M., Bertolino, S., Reeder, D. M., Bastos-Silveira, C., Sen, A., Sterner, B., Franz, N., Guidoti, M., Penev, L., and Agosti, D. Liberating host-virus knowledge from COVID-19 lockdown. EcoEvoRxiv – and a TWDG abstract on the same topic
-
Philosophy article: 2021. Sterner B., Elliott S., Upham N.S., Franz N.M. Bats, objectivity, and viral spillover risk. History & Philosophy of the Life Sciences 43:7
-
Taskforce public YouTube event: CETAF-DiSCCo Covid-19 Taskforce public event. Biodiversity knowledge hub for Covid-19 and preventing future pandemics. Link to YouTube-streamed talk (10 minutes).
-
Data publication of the initial host-virus data liberation: 2020. Poelen J, Upham N. S., Agosti D, Ruschel T, Guidoti M, Reeder D, et al. CETAF-DiSCCo/COVID19-TAF biodiversity-related knowledge hub working group: indexed biotic interactions and review summary. Zenodo. – Also this one on georeferencing horseshoe bat specimen records
-
-
Sept-Nov 2020: At the core this new work on mammal host-to-virus interactions is the reality that taxonomic names are the **keys** being used for both (i) identifying which host and virus organisms have been recorded to interact; and (ii) indexing the interaction evidence in global databases. Thus, linking taxonomic names to their associated meaning is critical to broadly linking biodiversity data into knowledge – “host-virus data” is just a special category of ecological interaction data, ultimately. If we are interested in the universe of ecological interactions, then we need to be much more rigorous about how we track & version the provenance of taxonomic naming schemes across datasets, researchers, and aggregated databases.
-
Our ASU group put some of these ideas into action for a panel discussion at TDWG2020 titled “Avenues into integration: communicating taxonomic intelligence from sender to recipient”, including the published abstracts listed there on The Automated Taxonomic Concept Reasoner and a case study looking at the dramatic shift from 2 to 25 recognized living species of Microcebus mouse lemurs: Redesigning the Trading Zone between Systematics and Conservation: Insights from Malagasy mouse lemur classifications, 1982 to present.
-
In parallel, I’ve been leading the Mammal Diversity Database into a new era of Github-based open-source databasing, thanks to a website re-design by our student web developer Schuyler Liphardt and the diligent efforts of our re-vamped MDD team. We now have 6,513 total species in the database and full versioning of the taxonomy on Zenodo and are in the process of curating a load of geographic, type specimen, and synonym data for release later this year. Generous support from the American Society of Mammalogists continues to move this project forward, supplemented by our ASU group now as well.
-
We applied for an NIH R21 grant aimed at broadly improving the public knowledge of mammal-to-virus interactions, for which I assembled a great multidisciplinary team from ASU (Beckett Sterner, Nico Franz, Arvind Varsani, and Jonathan Rees), Bucknell (DeeAnn Reeder), Univ of New Orleans (Atriya Sen), Plazi (Donat Agosti) and GloBI (Jorrit Poelen). This proposal (“Intelligently predicting viral spillover risks from bats and other wild mammals”) received strong scores, and is thus very likely to be funded starting in July 2021.
-
-
December 2020: I fully moved to Tempe AZ from my pandemic home in New Haven CT, allowing more regular access and engagement with the ASU Mammal Collections and NEON Biorepository.
-
Jan-Mar 2021: Together with Vertebrate Collections Manager Dakota Rowsey, we have started developing a project for sampling small mammals in the Madrean Sky Islands system of desert mountains. This work aims to better characterize the diversity, abundance, and genetic connectivity of mammal species and their pathogens, and use those patterns to query processes of community assembly and resilience to predicted climatic changes in the region. We plan to start work in the Santa Catalinas of SE Arizona and gradually work up to a broader comparative study across AZ, NM, and the Mexican states of Sonora and Chihuahua for which these Sky Island habitats extend.
Overall, I’m elated to be in Arizona, now teaching Evolution (BIO 345) to an online class of 170 ASU students and engaging with our growing group in the School of Life Sciences and Biodiversity Knowledge Integration Center (BioKIC). My work on Mammalia phylogeny, ecological causes of speciation, and comparisons to the genus-level fossil record also continues, though to do that justice will certainly need its own post :)
If you made it to the end here, then you deserve some sledding pandas! Here’s to a positive outlook for the rest of 2021…
Sincerely, –nate
