NIH R21 award outlined in The Lancet Planetary Health
Glad to share that I’m leading a new NIH R21 project to liberate mammal-to-virus data from publications – and then annotate them w/ supporting evidence to strengthen spillover risk models. Our approach is newly outlined in a Viewpoint for @TheLancetPlanet.
PDF / DOI: https://doi.org/10.1016/S2542-5196(21)00196-0
ASU news article: news.asu.edu/20211019-biolo…
Tweets: twitter.com/n8_upham/…

|
The grant “Intelligently predicting viral spillover risks from bats and other wild mammals” is based from ASU’s School of Life Sciences with co-I Beckett Sterner & team including Arvind Varsani, Nico Franz, Prashant Gupta, with Jorrit Poelen of GloBI & Donat Agosti of Plazi. DeeAnn Reeder is the other PI for the grant (based at Bucknell University), and has been critical to launching the study.
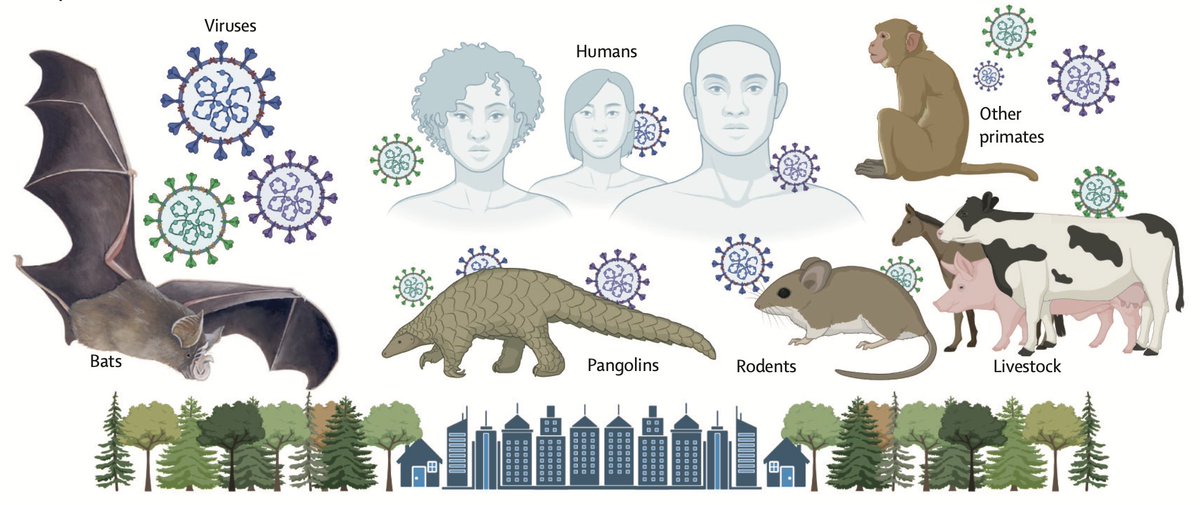
Goal: Improve existing host-virus data, which to date has largely been focused on viral biology rather than wild host ecology.
|
The problem is that existing host-virus data is often disconnected from evidence of either interaction (e.g., assay type, sample size) or taxonomy (e.g., voucher specimen, concept of species), which makes it hard to accurately model host-virus sharing w/ data collected long ago.
Our solution is two-part: (1) liberate #taxoknowledge from pubs w/ automated tools of Plazi.org & connect it w/ current taxonomy of the Mammal Diversity Database then (2) use the resulting taxonomic ‘passkey’ to search & unlock host-virus evidence in pubs for linking as digital knowledge.
We outline our taxonomy-centric approach in the @TheLancetPlanet article (doi.org/10.1016/S2542-…), emphasizing that the ‘unlocking dark data’ approach is inefficient vs. the ‘created connected’ publishing of Pensoft Publishers.
Digital knowledge needs to be the goal of all publishing.

|
Building taxonomic & interaction intelligence opens the door for creating probabilistic knowledge graphs of host-virus ecology, which is what’s needed to robustly predict viral spillover risks at finer levels than just “bats” or “rodents” (2/3rd of living mammal species)
Sharing the resulting improved host-virus data for continued re-use is the primary goal of this project — spearheaded by the efforts of GloBI to create lightweight, open source resources for the community: e.g., globalbioticinteractions.org/browse/?intera…
Grateful to the CETAF / DiSSCo Taskforce for initiating these efforts, including co-authors. Here is the full citation:
- Upham N.S., Poelen J.H., Paul D., Groom Q.J., Simmons N.B., Vanhove M.P.M., Bertolino S., Reeder D.M., Bastos-Silveira C., Sen A., Sterner B., Franz N.M., Guidoti M., Penev L., Agosti D. 2021. Liberating host–virus knowledge from biological dark data. The Lancet Planetary Health. https://doi.org/10.1016/S2542-5196(21)00196-0
