'Evolution of the largest mammalian genome' article is published in GBE
My collaborators and I published our first article on the genomics of Argentine vizcacha rats, which have 1.3x larger genomes than any other mammal (~8 Gb haploid, vs ~3 Gb mean for mammals).
^Evans, B. J., ^Upham, N. S., Golding, G. B., Ojeda, R. A., and Ojeda, A. A. 2017. Evolution of the largest mammalian genome. Genome Biology and Evolution 9: 1711-1724. (^ denotes co-corresponding authors).
This work started in Aug 2014 with an expedition to Mendoza Province, Argentina while I was a post-doc at McMaster University to collect high quality tissues from the red vizcacha rat, Tympanoctomys barrerae (see the Human Geographic article I wrote about that successful trip). We subsequently returned in Feb 2015 to collect matching tissues from Octomys mimax, the closest living relative of Tympanoctomys that has a normal-sized genome, to evaluate evidence for the hypotheses of Tympanoctomys genome expansion via mechanisms of whole genome duplication vs. repetitive element accumulation.
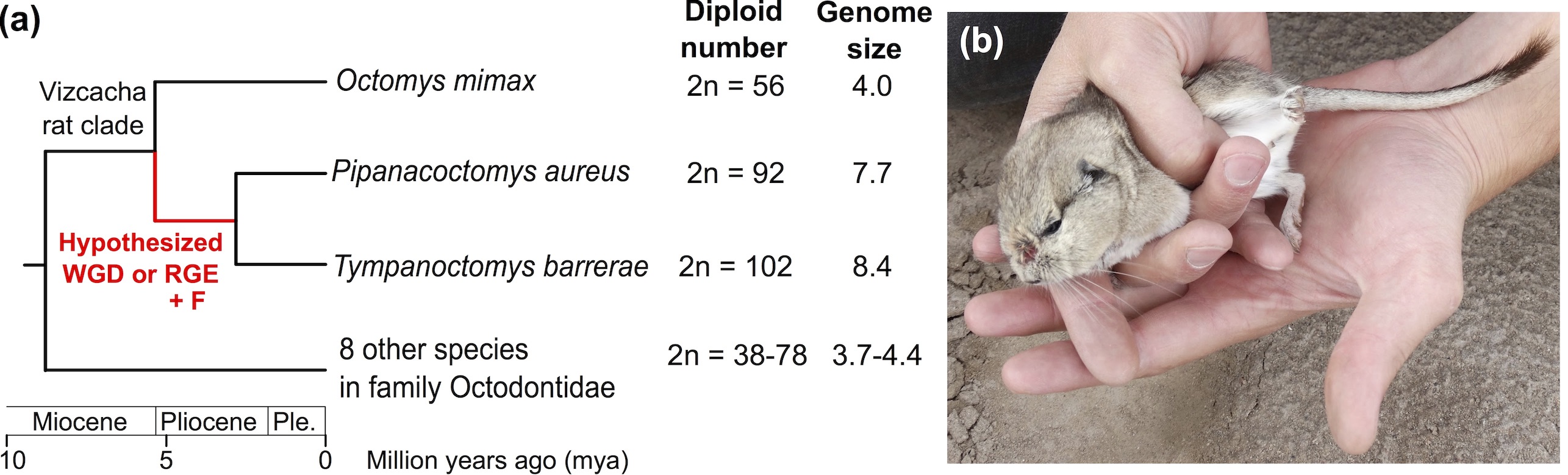
Results from RNAseq transcriptomes and HiSeqX short-reads found no evidence for widespread duplicated genes (expected if allopolyploidy had occurred) and instead uncovered abundant repetitive DNA. We conclude that repetitive elements such as retrotransposons are likely responsible for the rapid genome expansion of Tympanoctomys during the past 5 million years, but highlight that many questions remain. Foremost is why the 102-chromosome complement of Tympanoctomys is nearly all bi-armed, and how their chromosomal lability fits into the larger picture of Octodontidae and Octodontoidea evolution.
See this popular coverage of our article in ScienceDaily, on CBC Radio, and a great blog on The Molecular Ecologist.
Its an exciting story and we hope more to come!

